Looking back, 2025 was an exciting year for proteomics-driven discovery! A growing number of publications show how advanced sample preparation technologies are transforming the way we explore biology. From diving deeper into plasma, cerebrospinal fluid, and serum to uncovering the molecular mechanisms behind disease, these studies demonstrate the true power of high-quality proteome profiling. Thanks to robust, scalable workflows designed for clinical and translational research, these publications highlight how PreOmics solutions help scientists go further—delivering reproducible depth, sensitivity, and confidence in even the most complex biological systems.
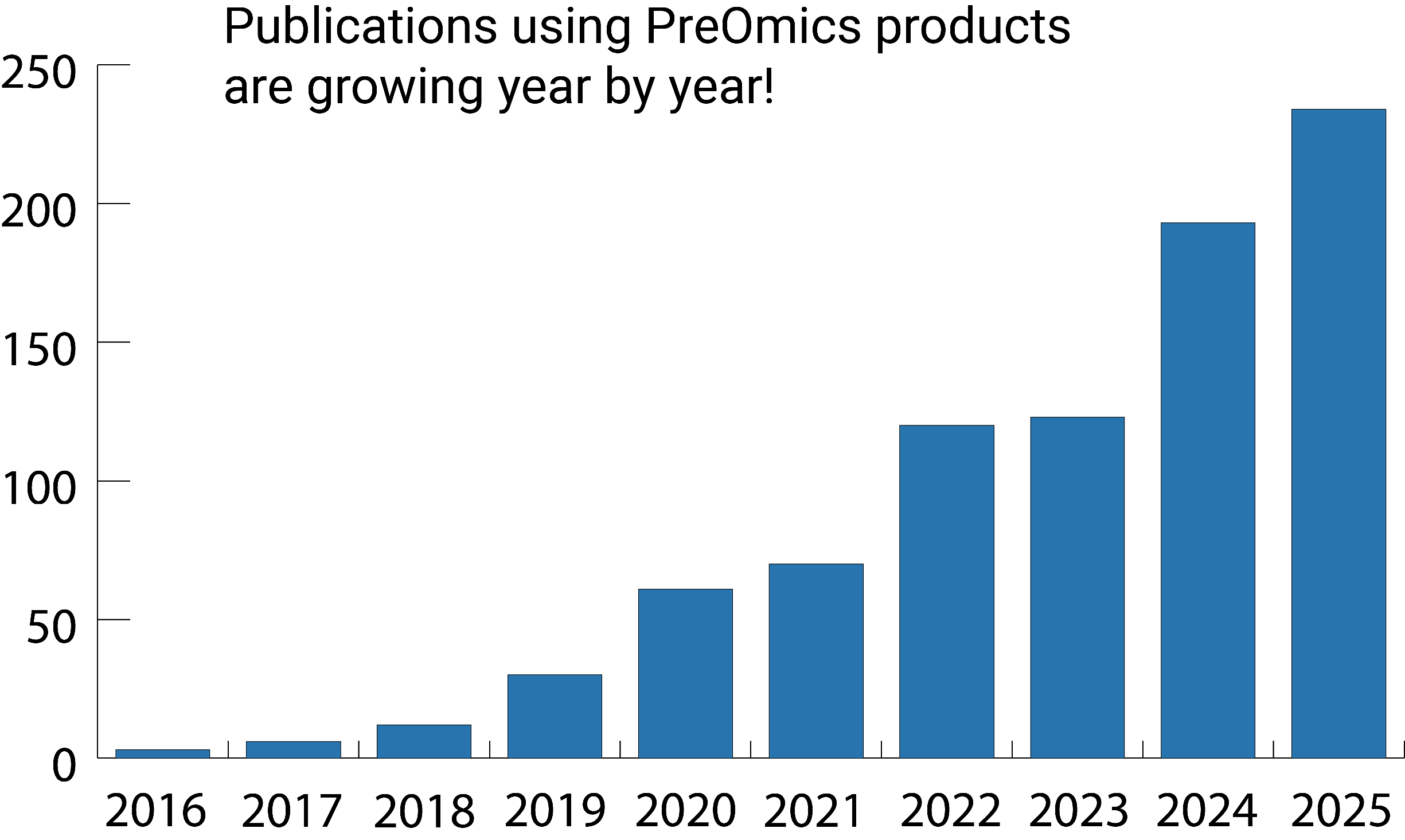
Check out our top-picked publications from 2025 just for you!
Muscle Physiology: Skeletal Muscle Proteomics Reveals Reversible Changes and Lasting “Memory”
Capturing dynamic changes in skeletal muscle proteomes across repeated training cycles is challenging due to tissue heterogeneity and the need for rapid, reproducible processing of multiple biopsies. Researchers employed BeatBox® for rapid tissue homogenization and iST for streamlined sample preparation, enabling quantitative comparison of acute and long-term proteomic changes. The integrated workflow enabled the detection of both reversible protein expression changes after individual training sessions and persistent “proteomic memory” signatures that remained after detraining. This approach revealed coordinated pathways involved in muscle adaptation, mitochondrial function, and metabolism, demonstrating how PreOmics solutions facilitate high-throughput studies of complex tissue samples.
Read publication: Hulmi, J.J. et al. Human skeletal muscle possesses both reversible proteomic signatures and a retained proteomic memory after repeated resistance training. The Journal of Physiology (2025).
Biomarker Discovery: Delving Deeper into the Human CSF Proteome
Quantifying low-abundance proteins in human cerebrospinal fluid (CSF) presents several challenges, such as the high dynamic range of protein concentrations, interference from abundant plasma proteins, and limited sample volumes. In a recent study published in the Journal of Proteomics, researchers applied an enrichment strategy using ENRICH-iST to address these issues. It enabled the enrichment of low-abundance proteins while maintaining high reproducibility and low coefficients of variation, providing deep proteome coverage from minimal CSF volumes. By streamlining sample handling and reducing variability, the ENRICH technology supports high-throughput biomarker discovery and clinical applications in neurological disease research.
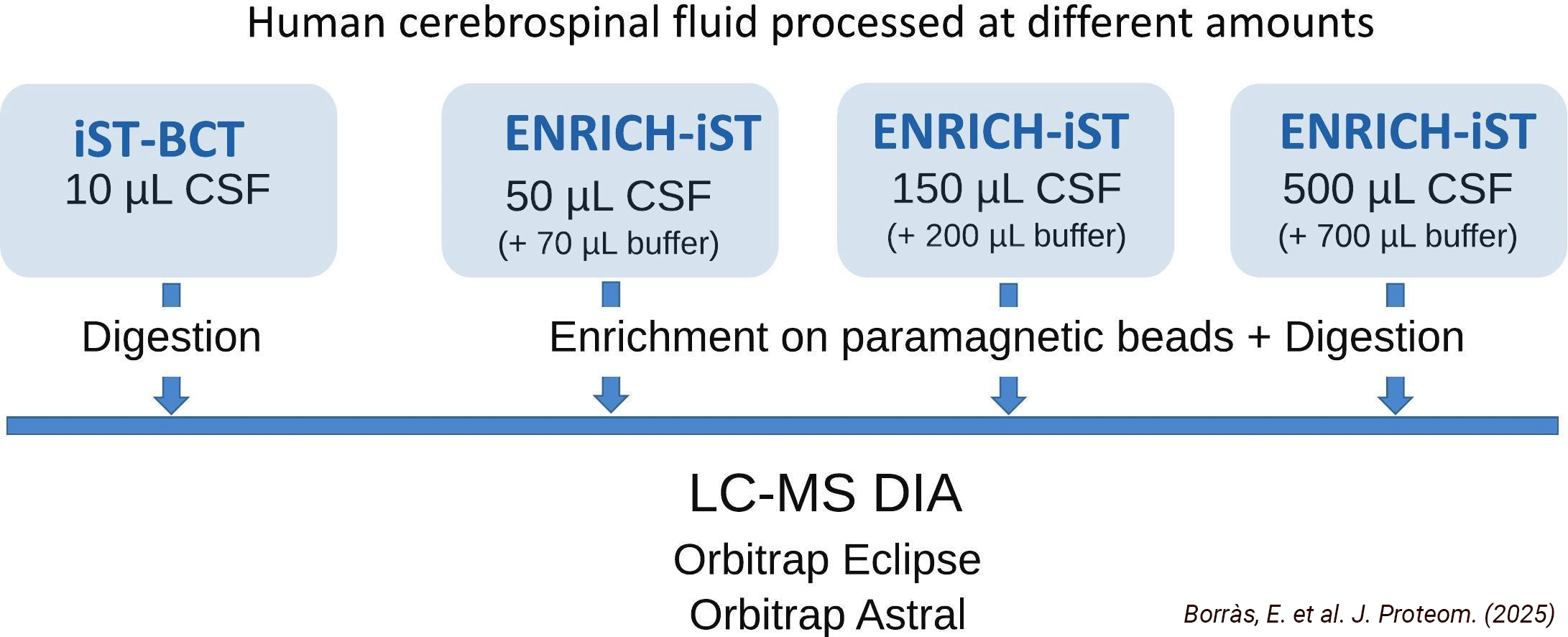
Read publication: Borràs, E. et al. Enhanced proteome profiling of human cerebrospinal fluid using a commercial plasma enrichment strategy. Journal of Proteomics (2025).
Neurodegenerative Diseases: β-Amyloid Sparks Microglial Signaling That Fuels Tau Pathology
Resolving microglial proteomic responses to β-amyloid requires sensitivity to low-abundance signaling proteins and reproducibility across multiple conditions. In this study, published in Molecular Neurodegeneration, researchers combined iST for standardized sample preparation and PepSep column for precise separation of peptides and proteins, enabling consistent quantification of GPC4, APOE, and downstream tau-associated pathways. The workflow minimized technical noise, allowing reliable mapping of microglial contributions to neuronal tau pathology and toxicity. These findings demonstrate how integrated proteomic workflows can uncover microglia-mediated mechanisms underlying tau pathology in neurodegenerative disease models.
Read publication: Holmes, B.B. et al. β-Amyloid induces microglial expression of GPC4 and APOE leading to increased neuronal tau pathology and toxicity. Molecular Neurodegeneration (2025).
Autoimmune Disease: Serum Fibrinogen Is Not a Biomarker for Myasthenia Gravis
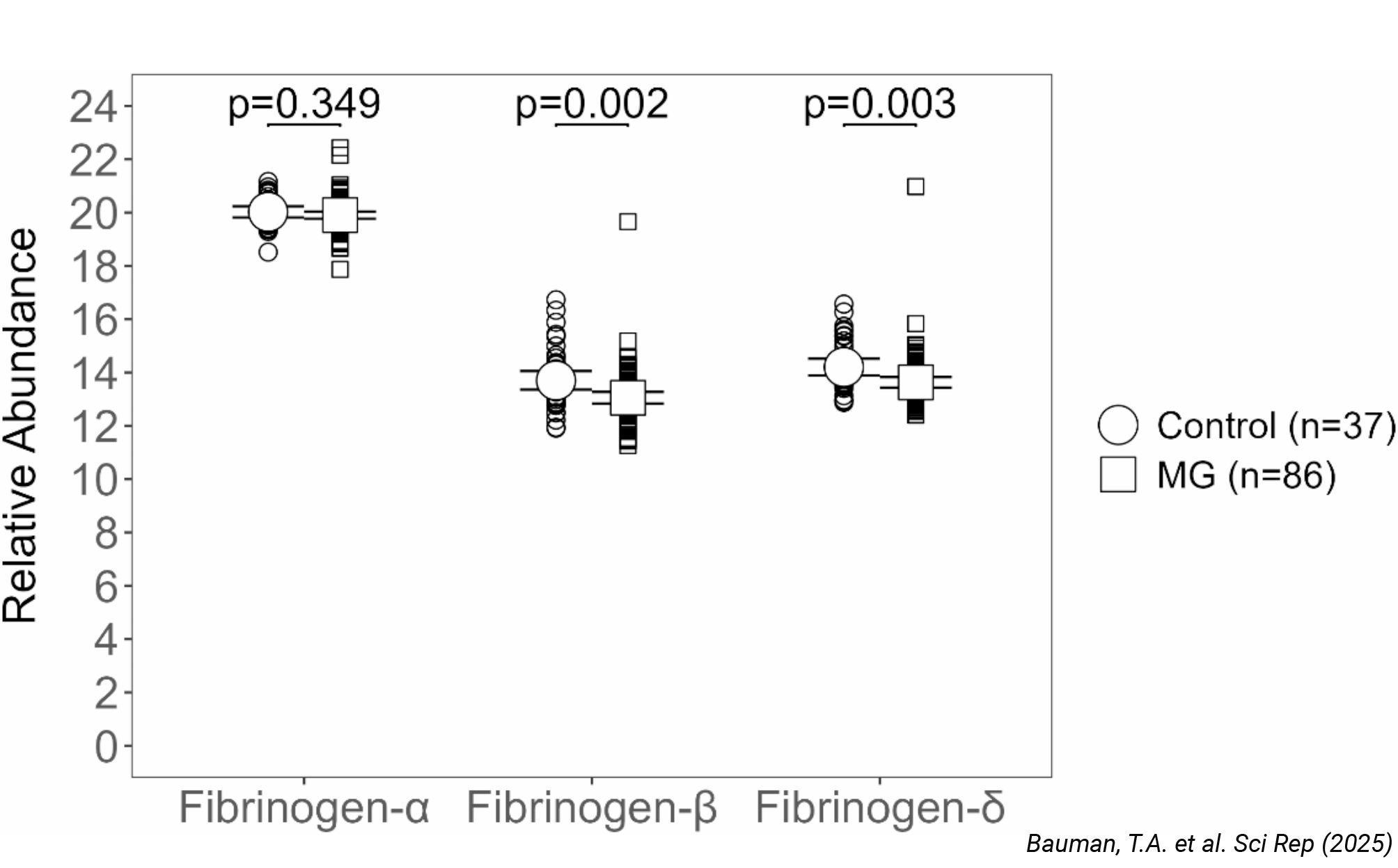
Detecting subtle proteomic differences in autoimmune disease requires workflows capable of handling sample complexity while maintaining high quantitative accuracy. Using ENRICH-iST for sample preparation and the PepSep column for peptide separation, researchers analyzed serum samples from myasthenia gravis patients. This approach minimized technical variability and interference from abundant serum proteins, demonstrating that fibrinogen levels remain unaltered in this patient population. The study, published in Scientific Reports, underscores how robust, streamlined workflows can help clarify biomarker relevance and guide disease monitoring strategies.
Read publication: Bauman, T.A. et al. Serum fibrinogen is not elevated in patients with myasthen. Scientific Reports (2025).
Cognitive Aging: Linking Primary Cilia and Autophagy to Neuronal Resilience
Neuron-specific proteome analysis in hippocampal tissue is constrained by tissue scarcity, cellular complexity, and the requirement to preserve subcellular integrity. Published in Nature Aging, this research combines BeatBox® homogenization with BeatBox Tissue Kit 24x for multi-sample preparation, enabling detailed profiling of hippocampal neurons. The study revealed a functional link between primary cilia and autophagy pathways critical for cognitive resilience. High reproducibility and efficient peptide recovery allowed confident quantification of signaling proteins and autophagy regulators, providing mechanistic insight into neuronal health and potential neuroprotective targets.
Read publication: Rivagorda, M. et al. A primary cilia–autophagy axis in hippocampal neurons is essential to maintain cognitive resilience. Nature Aging (2025).
Neurodegenerative Diseases: A GPX4 “Fin-Loop” Unlocks Protection Against Ferroptosis
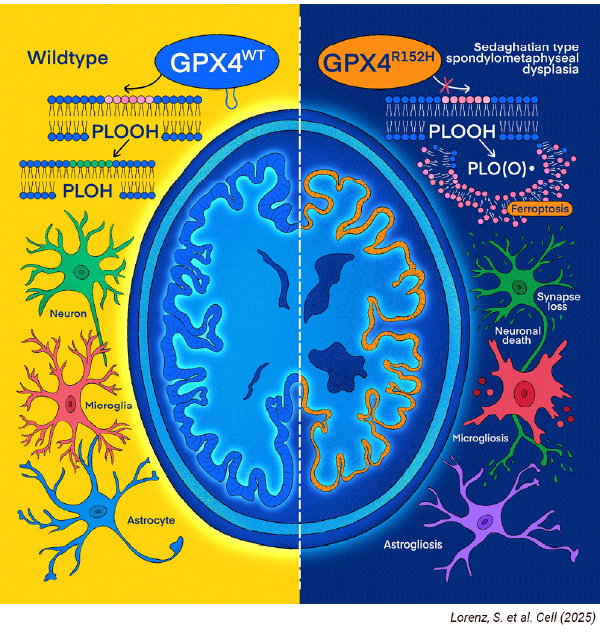
Investigating subtle structural-functional relationships in proteins like GPX4 requires workflows that preserve peptide integrity while maximizing coverage of low-abundance proteins. In this study, published in Cell, researchers used iST to process neuronal samples and capture the fin-loop-like structure responsible for GPX4-mediated protection against ferroptosis. The findings reveal direct in vivo evidence that ferroptosis plays a central role in neurodegeneration and can trigger a cell-nonautonomous inflammatory response. Although the study does not provide conclusive evidence for either the role of neuroinflammation as a driving force or as a consequence of neurodegeneration, it emphasizes the pathogenic nature of ferroptosis in the brain and underscores its promise as a therapeutic target across neurodegenerative diseases.
Read publication: Lorenz, S. et al. A fin-loop-like structure in GPX4 underlies neuroprotection from ferroptosis. Cell (2025).
Metabolic Disorders: Distinguishing Diabetes Types Through Renal Tubuli and Serum Proteomics
Differentiating type 2 diabetes mellitus (T2DM) from post-transplant diabetes mellitus (PTDM) at the molecular level is complex due to subtle proteomic differences and the need for high reproducibility across both tissue and serum samples. In this study, researchers analyzed microdissected proximal tubules and serum from kidney transplant recipients classified as normoglycemic (NG), pre-transplant T2DM, or PTDM. Key differences were identified in mitochondrial function and lipid metabolism pathways, which may contribute to the early development of diabetic nephropathy. Using ENRICH-iST, hundreds of serum proteins were consistently identified and quantified, revealing alterations in cholesterol and lipoprotein metabolism as well as potential biomarkers associated with each diabetes phenotype. These proteomic differences between PTDM and T2DM could support the development of targeted therapies and early diagnostic markers, ultimately improving transplant outcomes and patient management.
Read publication: Skandalou, E. et al. Proteome of renal tubuli and serum differentiate pre-existing type 2 diabetes and post-transplant diabetes in kidney transplant recipients. Proteomics Clinical Applications (2025).
These publications highlight how PreOmics technologies empower researchers to achieve reproducible, high-quality proteomic data, accelerating discoveries from fundamental biology to translational applications.
Looking for additional insights? Explore a curated selection of 2025 publications in proteomics and metabolomics from our Biognosys Group partners, Biognosys and biocrates.
Explore Our Full Publication Library



.webp)








